Introduction
Gastric cancer (GC) is one of the most prevalent cancers worldwide and remains a leading cause of cancer-related mortality, accounting for significant morbidity and healthcare burden globally [1]. Despite advances in diagnosis and therapeutic strategies, including surgical resection, chemotherapy, and targeted therapies, the prognosis for advanced GC patients remains poor, largely due to the development of drug resistance and recurrence [2]. Therefore, there is a need for novel therapeutic strategies targeting alternative cellular pathways to enhance treatment outcomes.
Suberoylanilide hydroxamic acid (SAHA), commonly known as vorinostat, is a histone deacetylase (HDAC) inhibitor that has demonstrated promising anticancer effects across a wide range of cancers [3]. Initially approved for the treatment of cutaneous T-cell lymphoma, SAHA exerts its anticancer activity through various mechanisms, including induction of apoptosis, cell cycle arrest, and disruption of mitochondrial functions [4–6]. Specifically, SAHA can trigger the intrinsic apoptotic pathway, modulate cell cycle-regulating proteins, and induce mitochondrial membrane potential (MMP) loss, culminating in cancer cell death.
In GC, epigenetic dysregulation, including altered histone acetylation patterns, has been recognized as an important contributor to cancer progression and therapeutic resistance [7]. HDAC inhibitors like SAHA are thus potential candidates to reverse these epigenetic abnormalities and suppress tumor growth. Although preliminary studies have indicated the potential efficacy of SAHA in solid tumors, including GC, its exact anticancer mechanisms in GC cells require further investigation [8].
Therefore, this study aimed to evaluate the anticancer activity of SAHA in GC cells and to elucidate underlying mechanisms involving apoptosis induction, mitochondrial dysfunction, cell cycle regulation, and the modulation of gene expression associated with nucleotide metabolism and cell cycle progression. Clarifying these mechanisms may provide important insights into SAHA’s therapeutic potential and guide future research and clinical application in GC management.
Methods
Four human GC cell lines (adenocarcinoma [AGS], KATO-III, MKN-45, and MKN-1) were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea). Each cell line was maintained in its recommended growth medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, under standard conditions (37°C, 5% CO2). Culture medium was refreshed every two to three days, and cells were subcultured once they reached approximately 70%–80% confluency.
Cells were seeded at a density conducive to logarithmic growth and allowed to adhere overnight. Subsequently, cells were treated with various concentrations of SAHA for 48 h. Following treatment, MTT solution was added to each well and incubated for 4 h as per the standard protocol. The formazan crystals formed were dissolved, and absorbance was measured using a microplate reader. Cell viability was calculated relative to untreated control samples.
For crystal violet staining, cells were seeded in multi-well plates, treated with SAHA for 48 h, and then fixed with a methanol-based fixative. Following fixation, cells were stained with a crystal violet solution for 30 min, after which excess dye was removed by gentle washing. The stained cells were air-dried, and intensity of staining was observed as an indicator of the adherent cell population. Quantification of the crystal violet signal was performed using ImageJ software.
To quantify apoptotic cell death, cells were exposed to SAHA for 48 h and then harvested by trypsinization. After centrifugation, cell pellets were resuspended in binding buffer containing Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI), following the manufacturer’s recommendations. Samples were incubated in the dark for 15 min, then immediately analyzed by flow cytometry to determine the proportion of cells in late apoptosis.
To assess MMP, cells were treated with SAHA for 24 h, harvested, and incubated with tetramethylrhodamine ethyl ester (TMRE) staining solution in the dark. After washing with a suitable assay buffer, cells were subjected to flow cytometric analysis. A reduction in TMRE fluorescence was interpreted as a loss of MMP, indicative of mitochondrial dysfunction.
Cells were seeded and treated with SAHA for 24 h. At the end of the treatment, cells were trypsinized, washed with ice-cold phosphate-buffered saline (PBS), and fixed in 70% ethanol at 4°C. The fixed cells were then resuspended in PBS containing ribonuclease A (RNase A) and stained with PI. The cell cycle distribution (subG1, G1, S, G2/M) was determined by flow cytometry, and data were analyzed to assess changes in cell cycle progression.
Total RNA was extracted from cells following 24 h of SAHA treatment using an AccuPrep® Universal RNA Extraction Kit (Bioneer, Daejeon, Korea). The extracted RNA was then reverse transcribed into cDNA using an AccuPower® RT PreMix (Bioneer) according to the manufacturer’s protocol. qPCR was performed using the AccuPower® 2X GreenStar qPCR Master Mix (Bioneer). The resultant cDNA was subsequently amplified in a real-time PCR system using AccuPower® 2X GreenStar qPCR Master Mix (Bioneer), along with specific primers for genes of interest and the housekeeping gene GAPDH. The sequences of the specific primers used in qPCR analysis are provided in Table 1. The 2−ΔΔCt method was employed to determine relative gene expression levels.
qPCR, quantitative real-time PCR; TYMS, thymidylate synthase; TYMP, thymidine phosphorylase; TK1, thymidine kinase 1; MTHFR, methylenetetrahydrofolate reductase; CCND1, cyclin D1; CCNE1, cyclin E1; CCNA2, cyclin A2; FOXM1, forkhead box M1; CHEK1, checkpoint kinase 1; MCM4, minichromosome maintenance protein 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
All experiments were conducted with at least three independent biological replicates. Data are presented as mean±SEM, where the SEM values were calculated from the variability across these independent replicates. Statistical comparisons between groups were performed using one-way analysis of variance (ANOVA) in the SAS statistical software, and p-values less than 0.05 were considered statistically significant.
Results
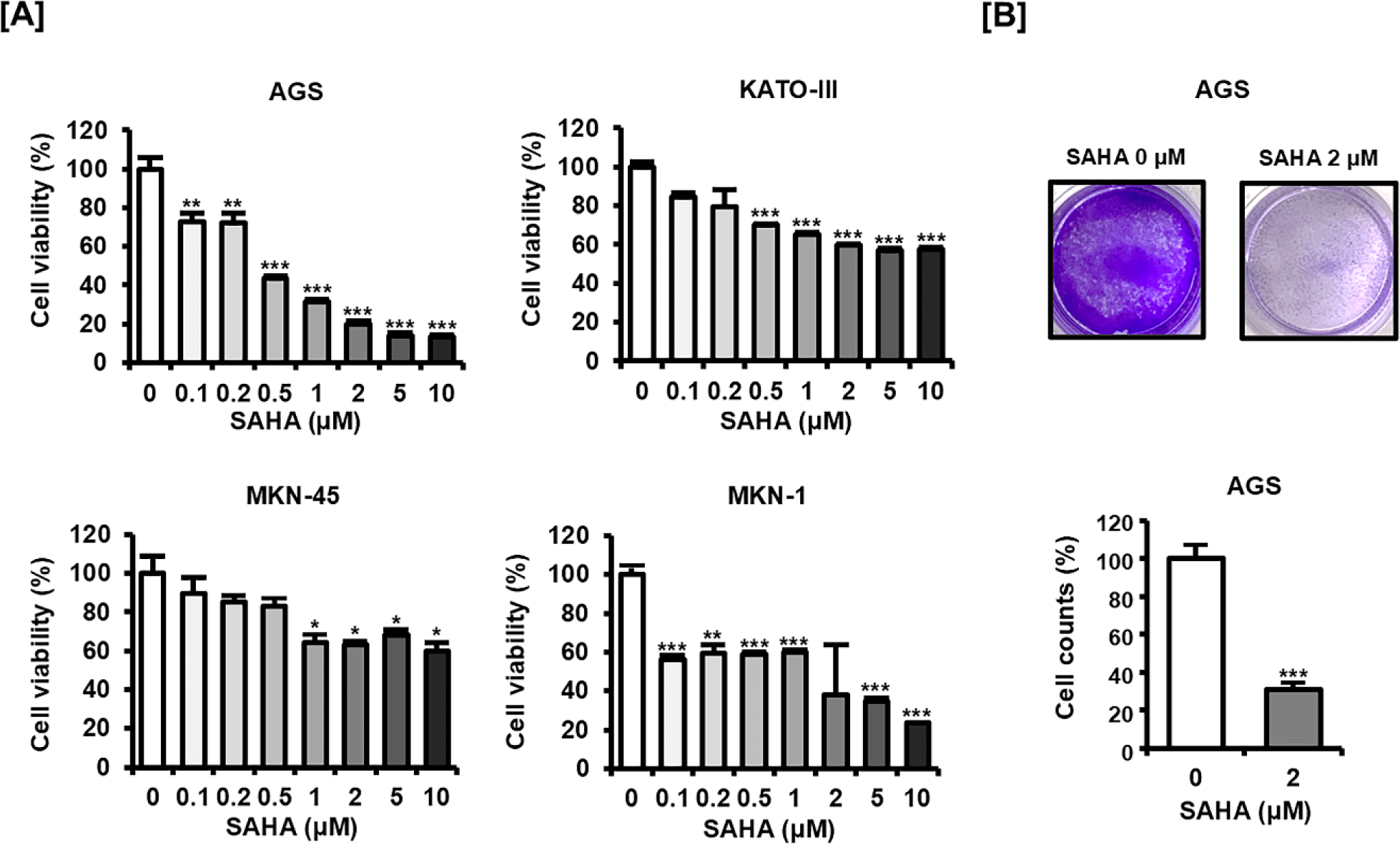
To explore the antiproliferative impact of SAHA in GC, four GC cell lines (AGS, KATO-III, MKN-45, and MKN-1) were treated with increasing concentrations of SAHA and assessed by MTT assay (Fig. 1A). All cell lines exhibited a dose-dependent reduction in viability, with AGS showing a particularly notable response. Given this pronounced sensitivity, AGS cells were specifically selected for subsequent detailed analyses to further characterize SAHA’s anticancer mechanisms in GC cells. Further experiments utilized AGS cells treated at a single concentration (2 μM). Consistent with the MTT results, crystal violet staining revealed that SAHA treatment led to a marked decrease in the density of AGS cell colonies (Fig. 1B). These observations illustrate that SAHA effectively suppresses GC cell proliferation and viability.
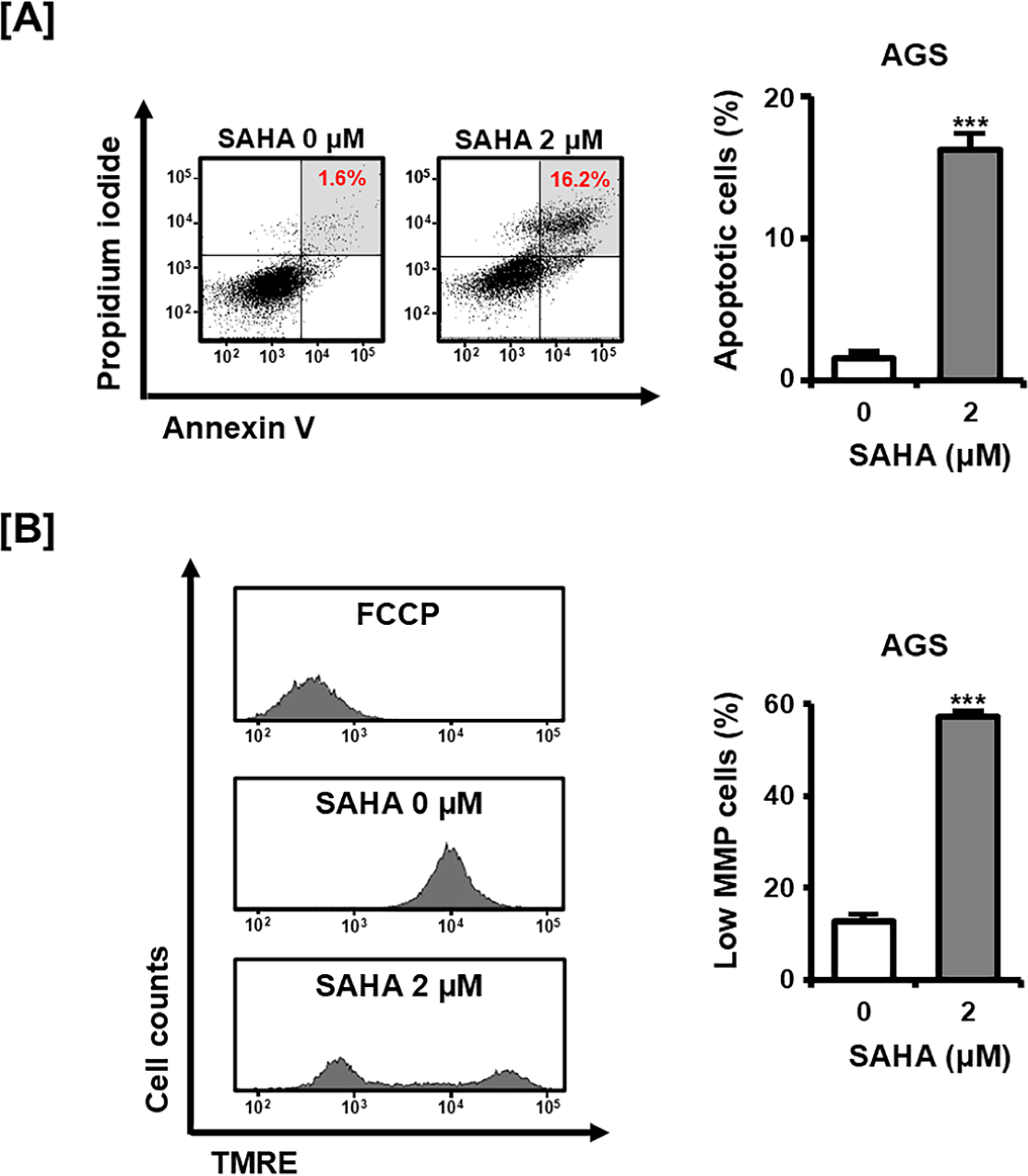
To determine whether the viability decrease was associated with apoptotic processes, AGS cells were subjected to Annexin V/PI staining (Fig. 2A). Following SAHA treatment, the proportion of double-positive (Annexin V+/PI+) cells, which represent late apoptotic cells, increased markedly compared with control cells. We next measured MMP using TMRE staining (Fig. 2B). SAHA-treated cells showed a substantial rise in the population with reduced fluorescence, indicative of mitochondrial depolarization. Collectively, these findings demonstrate that SAHA triggers apoptosis in GC cells, in part through mitochondrial dysfunction.
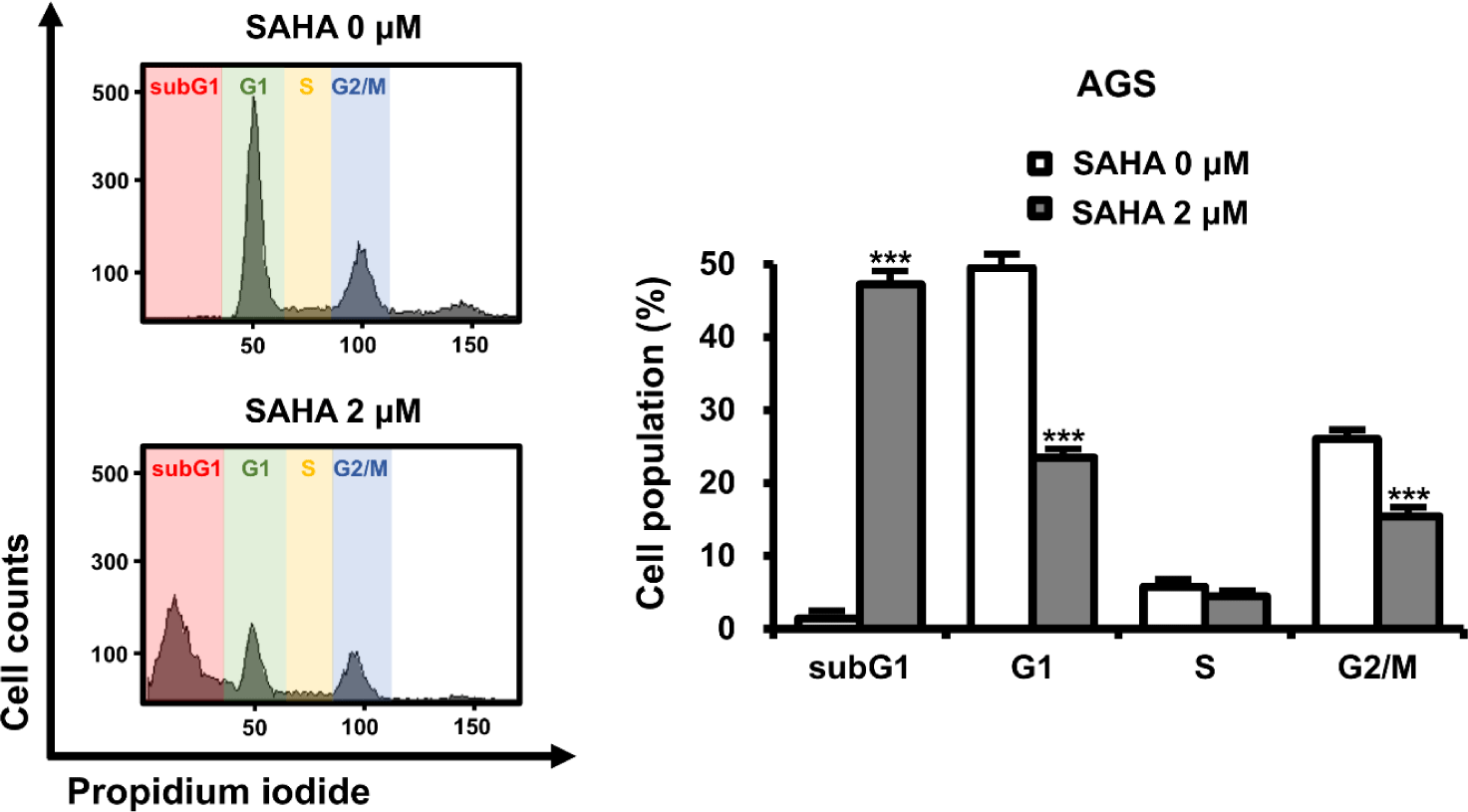
To investigate the mechanism underlying SAHA-mediated cell death, AGS cells were analyzed for changes in cell cycle progression via PI staining (Fig. 3). SAHA-treated cells exhibited an increased subG1 fraction, a hallmark of apoptotic cell death. Concomitantly, the percentage of cells in G1 and G2/M phases decreased. This shift in cell cycle profile aligns with the observed elevation in apoptotic cells, suggesting that SAHA-induced apoptosis is closely tied to cell cycle disruption in GC cells.
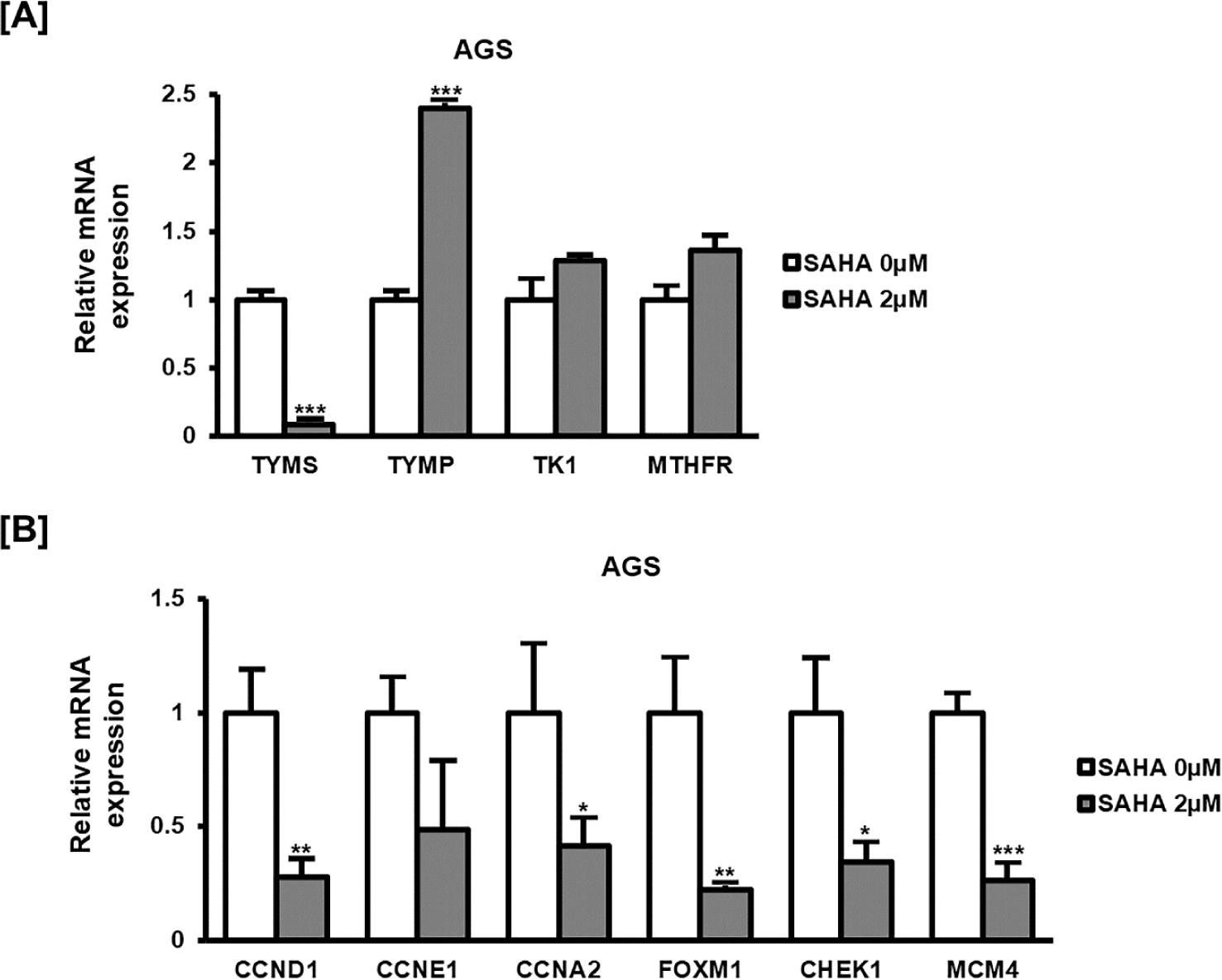
Finally, to gain insight into the molecular basis of SAHA’s antiproliferative effects, the mRNA levels of representative genes involved in nucleotide synthesis and cell cycle regulation were measured (Fig. 4). Thymidylate synthase (TYMS) expression was markedly decreased in SAHA-treated cells, while thymidine phosphorylase (TYMP) was substantially elevated. Thymidine kinase 1 (TK1) and methylenetetrahydrofolate reductase (MTHFR) showed minor variations. Similarly, genes critical for cell cycle progression, including cyclin D1 (CCND1), cyclin E1 (CCNE1), cyclin A2 (CCNA2), forkhead box M1 (FOXM1), checkpoint kinase 1 (CHEK1), and minichromosome maintenance protein 4 (MCM4), were downregulated. These results indicate that SAHA exerts its anticancer effects by influencing the expression of genes linked to DNA synthesis and cell cycle progression in GC cells.
Discussion
This study demonstrates the significant anticancer potential of SAHA in GC cells, providing insights into the molecular and cellular mechanisms underlying its antiproliferative effects. Our results show that SAHA markedly reduces GC cell viability through induction of apoptotic cell death, mitochondrial dysfunction, disruption of cell cycle progression, and modulation of key gene expression profiles related to nucleotide metabolism and cell cycle regulation.
The substantial increase in apoptosis and mitochondrial membrane depolarization observed in SAHA-treated AGS cells indicates that mitochondrial-mediated apoptosis is a central mechanism by which SAHA suppresses GC cell growth. These findings align well with previous studies suggesting that HDAC inhibitors, including SAHA, induce mitochondrial dysfunction and trigger intrinsic apoptotic pathways in various cancer types [9]. The observed increase in the subG1 population further reinforces the involvement of apoptosis as a primary response to SAHA, linking mitochondrial impairment directly to apoptotic induction.
The qPCR analysis revealed critical molecular changes following SAHA treatment, notably in genes involved in nucleotide metabolism. The significant downregulation of TYMS is particularly relevant, as TYMS is essential for DNA synthesis and repair. Reduced TYMS expression is known to impede tumor growth and is frequently targeted by chemotherapeutic agents like 5-fluorouracil (5-FU) [10,11]. Thus, SAHA-induced TYMS suppression could represent a novel mechanism contributing to its antiproliferative effects and potentially increasing sensitivity to DNA-targeting chemotherapeutics.
Conversely, the pronounced increase in TYMP expression upon SAHA treatment is notable given its dual roles in nucleotide metabolism and angiogenesis regulation [12,13]. Elevated TYMP levels may impact tumor growth and sensitivity to therapy by enhancing nucleotide salvage pathways and modulating the tumor microenvironment [14]. Although the exact implications of increased TYMP expression require further investigation, it may reflect compensatory metabolic adaptations triggered by SAHA-mediated nucleotide synthesis inhibition.
Additionally, SAHA’s impact on cell cycle-regulatory genes, including decreased expression of cyclins (CCND1, CCNE1, CCNA2), FOXM1, CHEK1, and MCM4, highlights its broad effects on tumor cell proliferation machinery [15,16]. Such transcriptional changes corroborate the observed disruption of normal cell cycle progression and reinforce SAHA’s potential to inhibit tumor growth effectively.
The results of this study highlight the promising clinical potential of SAHA in gastric cancer therapy. Given that SAHA is already FDA-approved for treating cutaneous T-cell lymphoma, it has well-established pharmacological characteristics and clinical safety profiles, facilitating potential translation to gastric cancer treatment [4]. However, comprehensive clinical studies are necessary to further define the optimal clinical use, therapeutic window, and long-term benefits of SAHA in managing gastric cancer.
Taken together, our findings provide a deeper understanding of SAHA’s multifaceted anticancer activity in GC. Future research should explore these gene-expression changes in clinical samples and evaluate potential synergistic effects of SAHA combined with conventional GC therapies. This study supports the continued investigation of SAHA as a promising candidate for GC treatment.
